problem of exporting the atomic model of DNA to PDB
-
When exporting an atomic DNA model to PDB for use in other programs (e.g., Schrodinger Suite), ATOM records are marked as HETATM, and residues have the non-standard name N A, which causes import problems. How can I correctly export a full-atom PDB that is compatible with standard molecular software?
 Example: AUTHOR GENERATED BY SAMSON 2025 R2 (8.0.0)
Example: AUTHOR GENERATED BY SAMSON 2025 R2 (8.0.0)
MODEL 1
HETATM 1 P N A 148.476-407.921 -26.809 0.00 P
HETATM 2 OP1 N A 148.722-409.286 -26.289 0.00 O
HETATM 3 OP2 N A 149.239-406.836 -26.153 0.00 O
HETATM 4 O5' N A 148,735-407,888 -28,387 0.00 O
HETATM 5 C5' N A 147,743-408,444 -29,270 0.00 C
HETATM 6 C4' N A 148,403-408,954 -30,537 0.00 C
HETATM 7 O4' N A 148,519-407,853 -31,484 0.00 O
HETATM 8 C3' N A 149,833-409,471 -30,379 0.00 C
HETATM 9 O3' N A 150,178-410,357 -31,440 0.00 O
HETATM 10 C2' N A 150,670-408,202 -30,528 0.00 C
HETATM 11 C1' N A 149,866-407,411 -31,556 0.00 C
HETATM 12 N9 N A 149,881-405,940 -31,325 0.00 N
HETATM 13 C8 N A 149,897-405,249 -30,135 0.00 C
HETATM 14 N7 N A 149,908-403,959 -30,275 0.00 N
HETATM 15 C5 N A 149,897-403,771 -31,650 0.00 C
HETATM 16 C6 N A 149,899-402,616 -32,448 0.00 C
HETATM 17 N6 N A 149,915-401,372 -31,948 0.00 N
HETATM 18 N1 N A 149,885-402,785 -33,783 0.00 N
HETATM 19 C2 N A 149,870-404,023 -34,273 0.00 C
CONECT 444 442
CONECT 445 442 446
CONECT 446 445 447
CONECT 447 446 448 449
CONECT 448 447 452
CONECT 449 447 450 451
CONECT 450 449
CONECT 451 449 452
CONECT 452 448 451 453
CONECT 453 452 454 462
CONECT 454 453 455
CONECT 455 454 456
CONECT 456 455 457 462
CONECT 457 456 458 459
CONECT 458 457
CONECT 459 457 460
CONECT 460 459 461
CONECT 461 460 462
CONECT 462 453 456 461
MASTER 0 0 0 0 0 0 0 0 473 2 462 0
END -
Dear @Dominic ,
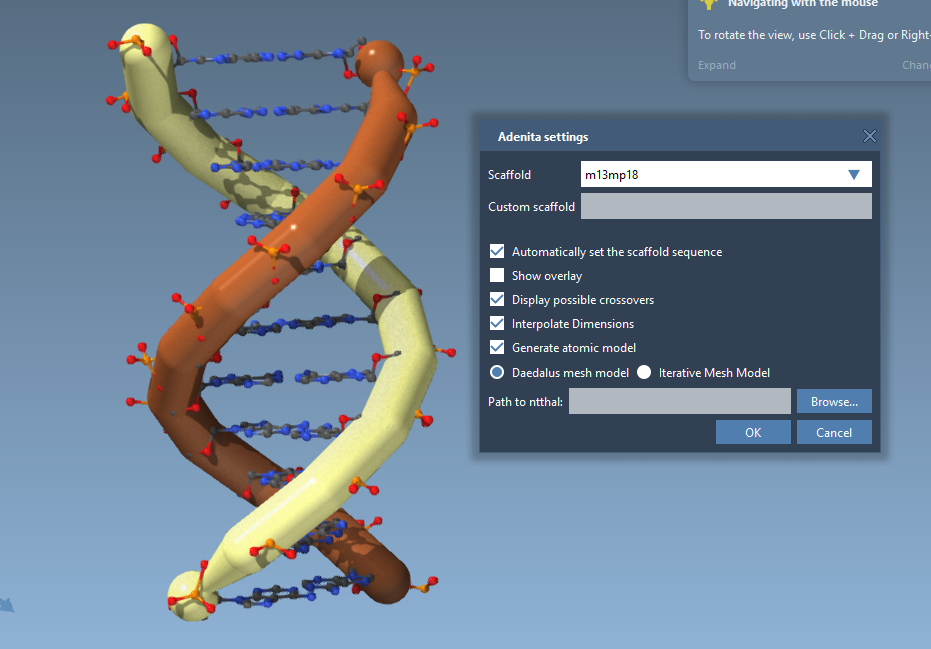
The "Automatically set the scaffold sequence" option in the settings is applied only when creating scaffolds, e.g., using the Wireframe editor.
It is not done for the DNA Strand Creator, so for structures done with it, you need to select them and click Generate sequence to add a random sequence to them before adding atoms:

Then the exported PDB file will refer to A, C, G, T names (but not DA, DC, DG, DT), as set by Adenita.
Please note that an exported PDB file might contain some dummy HETATM used by Adenita when building/connecting structures. You can remove them either in the PDB file itself or, if you no longer need to use them with Adenita, then directly in SAMSON after the atomic model has been generated via Select > Atoms > HETATM and then delete the selected atoms.
-