Facing problem in using AutoDock Vina
-
Recently I have added AutoDock Vina feature.....But I am facing little difficulty in using that app....Can you please provide me with a tutorial of how to dock ligands with a proteins...how to set grid points etc.
-
Hello there, the typical procedure is as follows:
- Select the protein (in the viewport or in the document view), then press the button "Set receptor".
- If you want flexible side-chains, select them (side-chain nodes in the document view, c.f. the Quick Start Guide, then press "Set flexible side-chains".
- Select the ligand (in the viewport or the document view), then press the button "Set ligand".
- If you have flexible side-chains, or a flexible ligand, you will see possible rotatable bonds in the viewport represented by green cylinders superimposed with bonds:
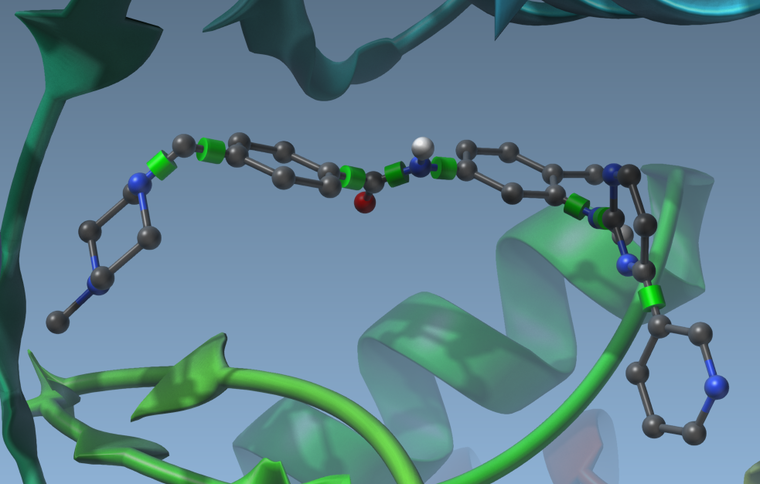
Using the delegate editor you can activate or deactivate these rotatable bonds by clicking on the cylinders. They will change colors when you do. Green means the bond can rotate, and red means the bond cannot rotate:
you can activate or deactivate these rotatable bonds by clicking on the cylinders. They will change colors when you do. Green means the bond can rotate, and red means the bond cannot rotate:
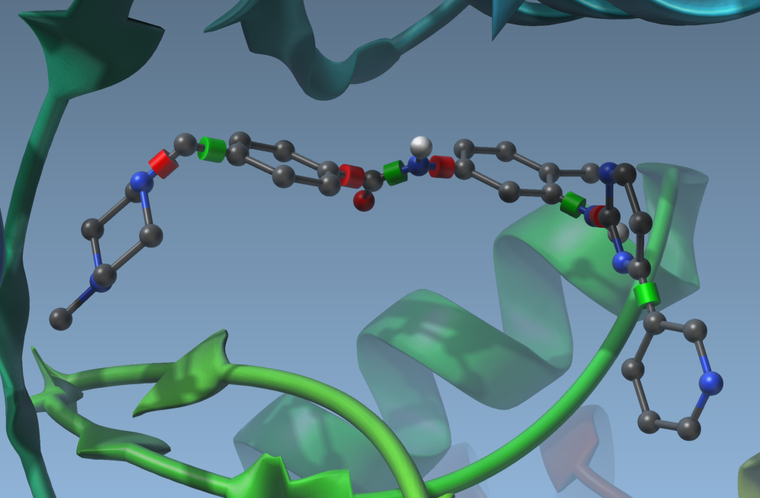
Having rotatable bonds makes the search slower but may result in more realistic docking. - Set the search domain by changing the grid center and size. You can do this either by changing the numbers in the App GUI, or by using the delegate editor and drag the yellow grid corners in the viewport.
- Set the Exhaustiveness parameter (the higher the better, but the longer the search).
- Set the Modes parameter (the maximum number of modes returned by the search algorithm).
- Press dock and wait for the results.
- Select which mode to display by using the combobox in the App. You can also restore conformations by double-clicking them in the viewport:
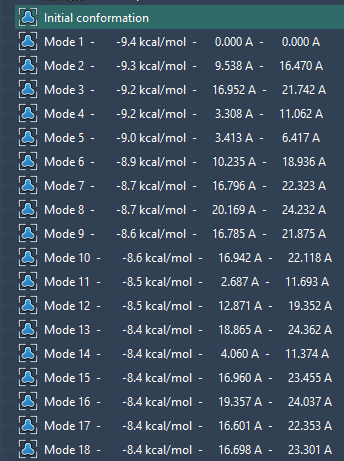
Hope this helps,
Stephane
-
Thank you so much sir....It really helped.