Ligand Receptor Interaction
-
Dear @HUMERA ,
To visualize hydrogen bonds you can use the Hydrogen Bond Finder element - just click Add and it will appear in you SAMSON installation next time you launch SAMSON.
-
@humera said in Ligand Receptor Interaction:
One more thing is that when I expand selection to 'side chains' within 6 angstrom of selected ligand, and then apply Licorice visual model. It displays few atoms as well along with side chains.
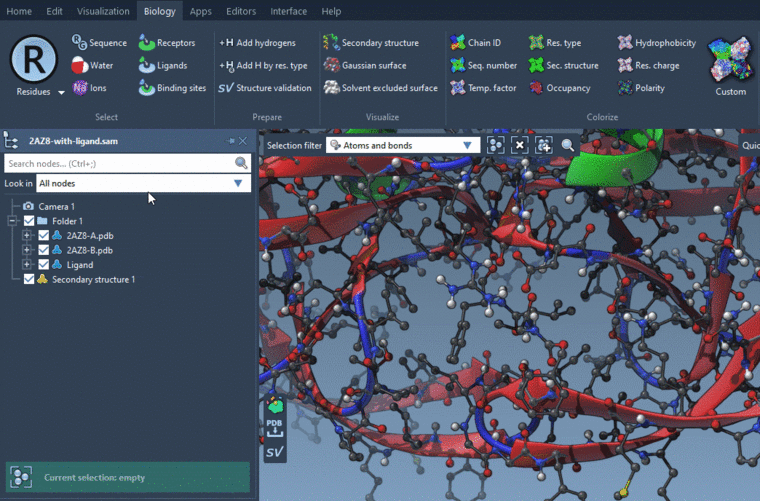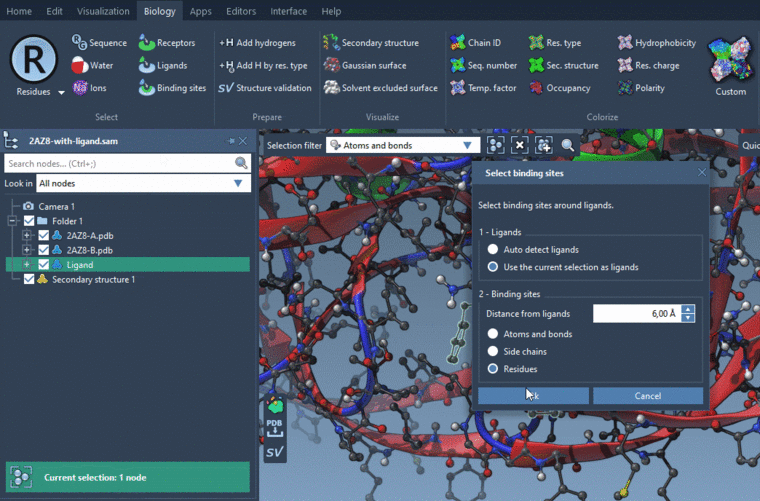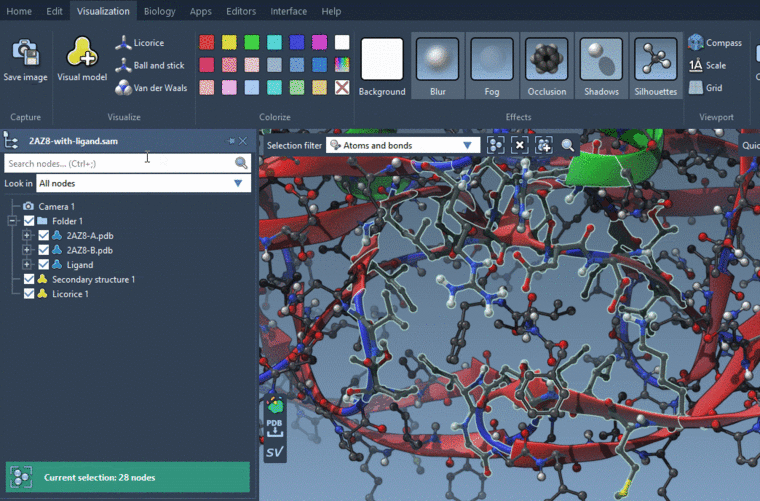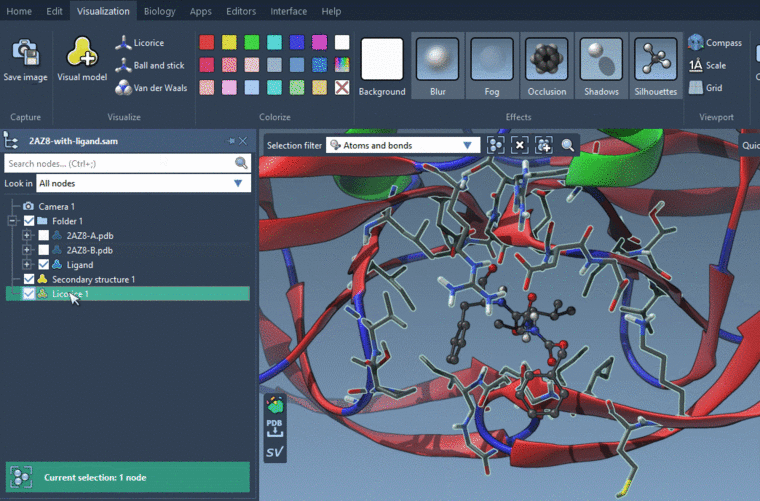
In short I want to display the side chains which are interacting with ligand, or which act as a binding site.To select side chains around a ligand:
- Select a ligand
- Biology menu > Binding sites: Select Use the current selection as ligands in the Ligands box and Side chains in the Binding sites box, and specify the distance from ligands.
To visualize them using the Licorice visual model: Visualization menu > Licorice (Ctrl+Shift+V). But I would recommend visualizing the whole residues with the licorice visual model. If you don't want to see atoms, you will need to hide them: uncheck nodes in the Document view which you want to hide.
-
Please see the gif attached:
-
Dear @dmitriymarin,
Thank you for your quick response.
I followed these steps but still certain atoms appear, and those atoms were hydrogen atoms. I resolved this issue by removing all hydrogen atoms from structure. But it is not a good solution since sometimes we have to display hydrogen atoms to indicate hydrogen bond.
Regards,
Humera -
I got it. Thank you.
Is there any SAMSON element available which can display cysteine disulphide bridges? -
Is there any SAMSON element available which can display cysteine disulphide bridges?
Cysteine disulfide bridges are displayed as bonds.
You cand find SS-bonds using e.g. Node Specification Language (NSL): S linking S (which stands for Sulfur atom linked to another Sulfur atom).
And to find precisely Cysteine disulfide bridges: (S in residue.type CYS) linking (S in residue.type CYS) (short version: (S in r.t CYS) l (S in r.t CYS)).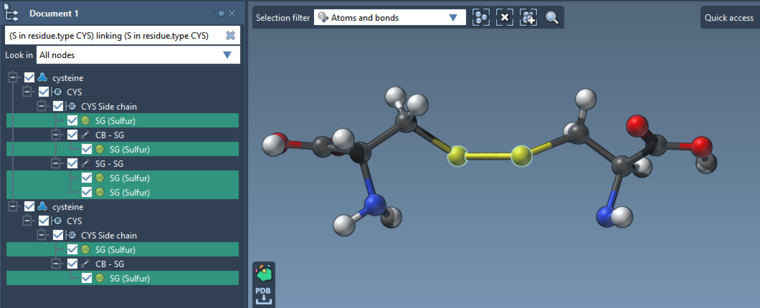
-
I followed these steps but still certain atoms appear, and those atoms were hydrogen atoms. I resolved this issue by removing all hydrogen atoms from structure. But it is not a good solution since sometimes we have to display hydrogen atoms to indicate hydrogen bond.
The hydrogen atom might appear if, for example, you add them yourself using e.g. Add hydrogens. You don't need to delete atoms to hide them, you can just hide the structure in e.g. Document view (see the gif in the answer above). To learn more, I recommend you to go through interactive tutorials in SAMSON: Help > Tutorials.
-
@dmitriymarin Got it. Thank you.
-
How can I show H-bonds at the interface between two proteins?
I tried selecting the binding site and then enlarging the selection, but the H bond finder detect all possible H-bonds, both intra-chain and inter-chain in the selection.
Thanks in advance -
Dear @Mariangela ,
To find and show hydrogen bonds you can use the Hydrogen Bond Finder Extension. It allows you to find all H-bonds in the selected system or between two systems, for example, between a receptor and a ligand.
To find/show H-bonds between a receptor and a ligand, perform the following steps (see the gif at the end):
- in the Hydrogen Bond Finder Extension click on "...in the current selection and the system";
- select the receptor in the Document view and in the Hydrogen Bond Finder click Set;
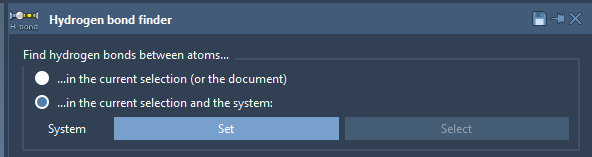
- set the Settings of the Hydrogen Bond Finder if necessary;
- now, select only the ligand in the Document view and in the Hydrogen Bond Finder click Add hydrogen bond visual model
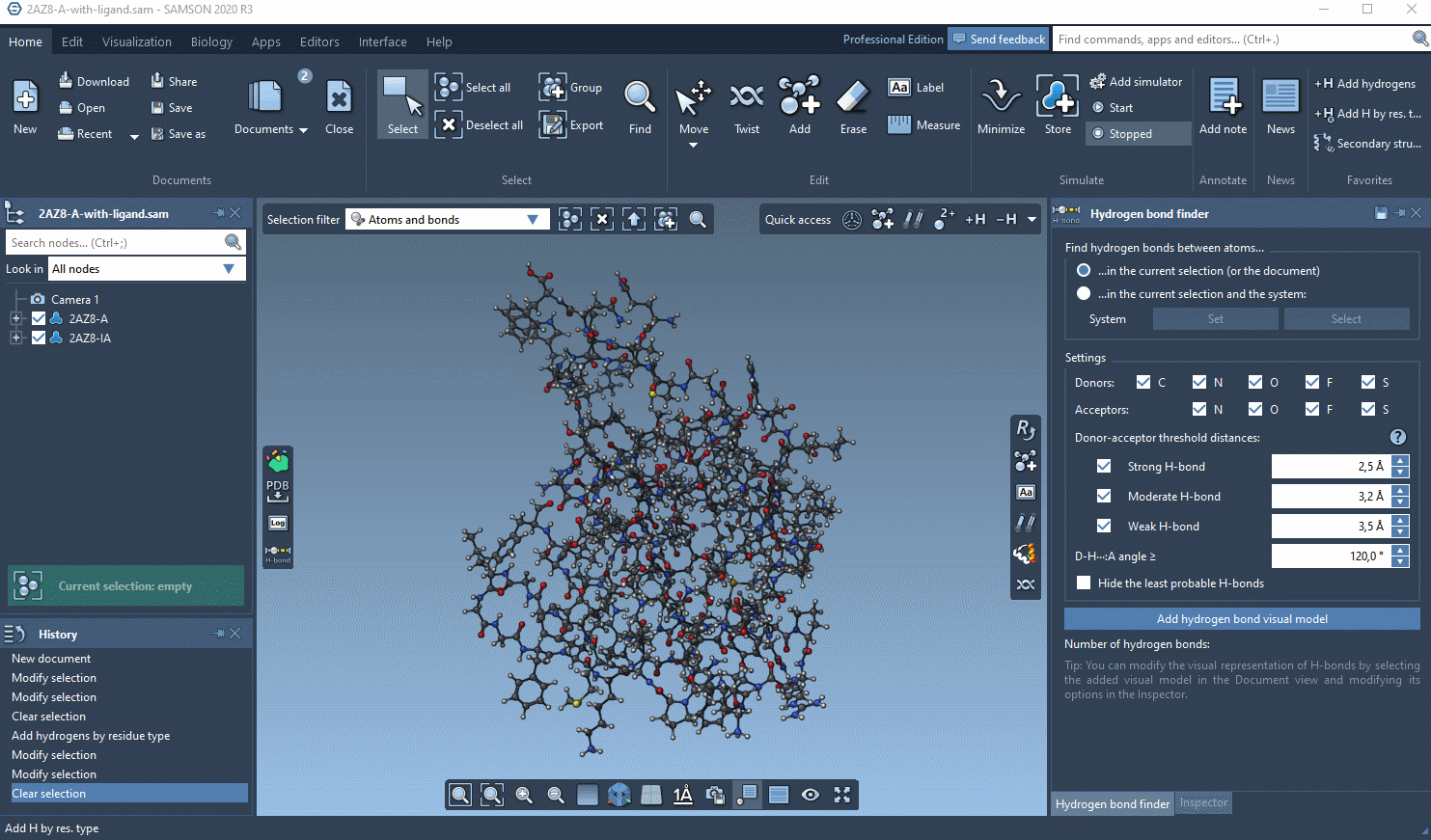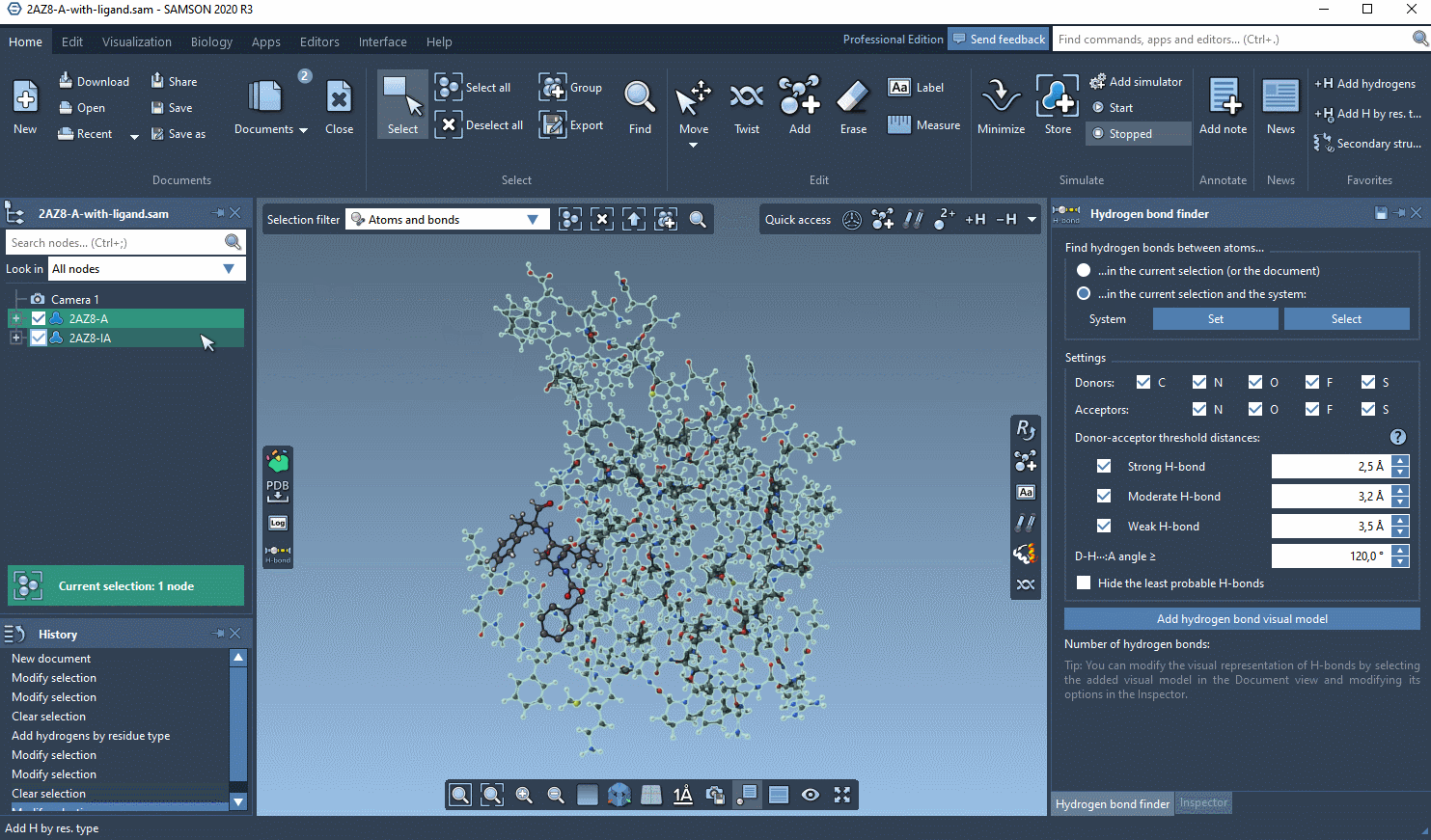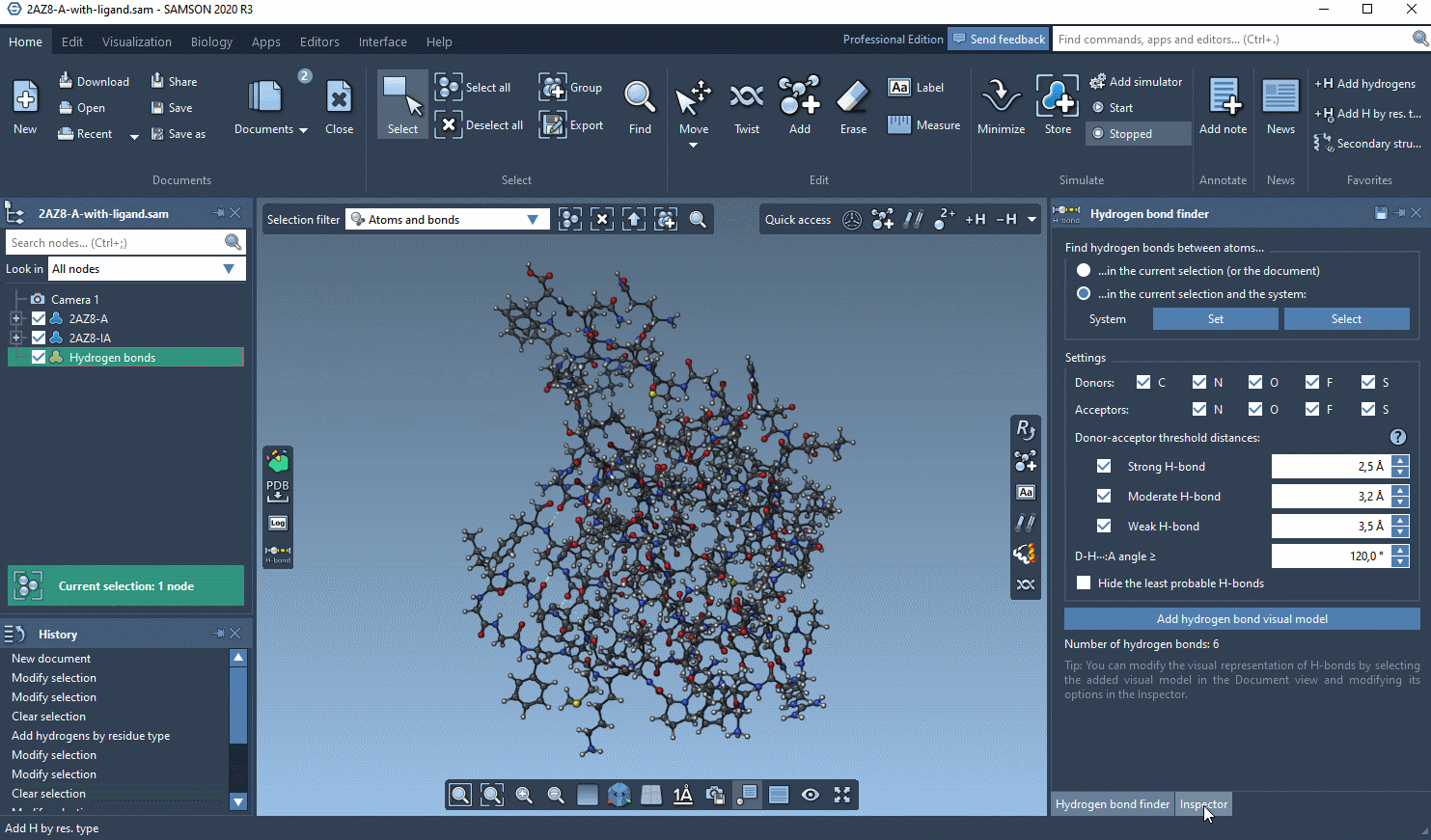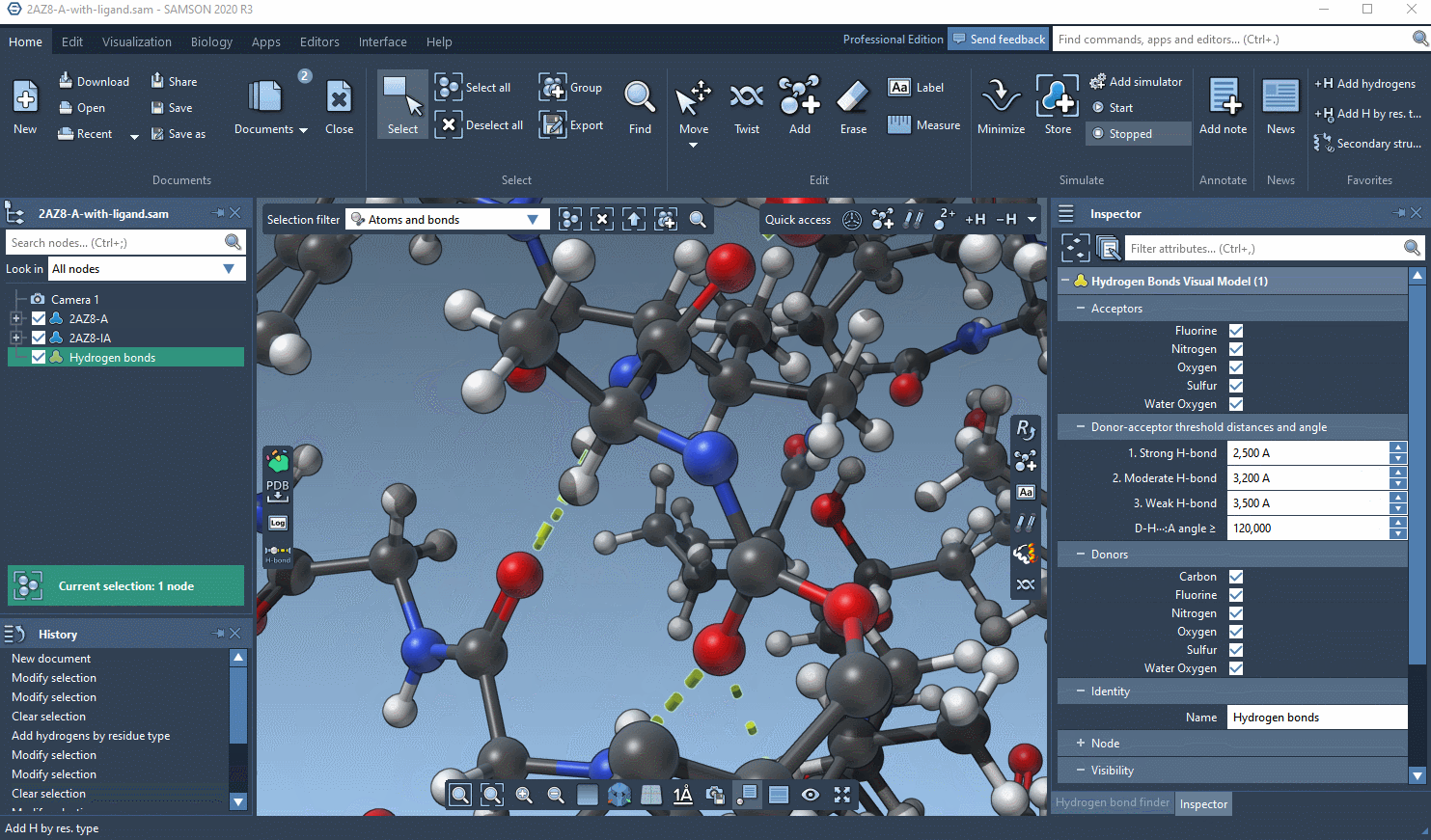
Now you should have a new visual model in the Document view. If you select it, you can modify its options in the Inspector.
See an example below: